

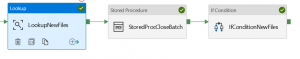
If you’ve ever found yourself faced with two copies of what is meant to be the same directory, then rsync can synchronize them into one directory containing the latest changes from each. What does make a difference even locally, though, is the command’s ability to differentiate files that share the same name but which contain different data. Locally, that fact doesn’t necessarily mean much, but if you’re copying gigabytes of data to a remote server, this feature makes a world of difference. I am trying to recurse through a directory and copy it from A to B. If you use rsync to copy a file into a directory, but that file already exists in that directory, then rsync doesn’t bother performing the copy operation. The command’s true power lies in its ability to not copy when it’s not necessary. There are many regex guides online – I often head over to Rubular to test a pattern if I’m having trouble (note that it’s actually designed for Ruby and not R but they seem similar enough that it has always worked so far). The drive contains around 800gb of data with over a million files that are 20-80K each. The only tricky bit can be getting the regex pattern right to pull out the files you want to manipulate. Im using robocopy to copy files to a samba share. Select BatchDate, DATEADD (HH, -2, BatchDate) AS BatchDate2 From BatchControl Where BatchId (Select Max (BatchId) From BatchControl Where BatchName GAVFiles AND Completed 1) Then I create new row into BatchControl table. This will chug away for a bit (time for a coffee, anyone?) and then produce a vector of TRUE/FALSE the same length as your list.of.files that identifies whether it was able to copy them or not. First I use Lookup activity to get previous executions max file creation date.

List.of.files <- list.files(current.folder, "SDM.tif$") New.folder <- "H:/Where I want my files to be copied to"
# identify the foldersĬurrent.folder <- "C:/Where my files currently live" In my case, I wanted to identify all Geo-tiff formatted rasters whose filenames ended in “SDM” and copy them to a new directory. Basically, you point R at the directory where your files live, identify the files that you want to manipulate and then tell it what you want to do to them. R has a nice set of file manipulation commands in the base package that make it really easy to find out if a file exists ( file.exist), rename files ( file.rename), copy them to a different directory ( py) or delete them ( lete). So enter my foray into copying files using R. Not only would this have been incredibly tedious(!), it’s highly likely that I would have made a mistake and missed something important or copied over files that they didn’t need. This is purely matching filename only and not doing a binary compare. While this is not typically a difficult task to do manually, I didn’t want to copy all of the files within the folder and there was no way to sort the folder in a sensible manner that meant I could select out the files that I wanted without individually clicking on all 723 files (out of ~4,300) and copying them over. If the file is new, then copy it, if its already in the destination folder, ignore it. Following on from my recent experience with deleting files using R, I found myself needing to copy a large number of raster files from a folder on my computer to a USB drive so that I could post them to a colleague (yes, snail mail – how old and antiquated!).


 0 kommentar(er)
0 kommentar(er)
